

Site-directed mutagenesis is a technique conceived in the 70s used to mutate specific DNA sequences in vitro. It relies on synthetic short single-stranded DNA fragments, or oligonucleotides, that contain designated mutations to act as templates in the presence of DNA polymerase enzyme.
In the late 80s, Polymerase Chain Reaction (PCR), another laboratory technique, was incorporated into site-directed mutagenesis workflow. The modified site-directed mutagenesis is a simple and efficient technique that has rapidly become the cornerstone of gene and protein function studies.
As a result, Michael Smith, the inventor of site-directed mutagenesis, and Kary B. Mullis, the inventor of PCR, were awarded the equally shared 1993 Nobel Prize in Chemistry.
Originally termed oligonucleotide-directed mutagenesis, the technique was primarily developed in a single-stranded DNA Escherichia coli phage ΦX174. The core idea of the technique stemmed from the following discoveries in the bacteriophage:
These discoveries inspired Smith to develop an oligonucleotide-based mutagenic method. In this method, a short strand of synthetic oligonucleotides contains the predefined mutation and acts as a mutagen that alters a specific nucleobase on the specified DNA position. The first site-directed mutagenesis reported by Smith and his team used one strand of oligonucleotides as mutagens to prime with a single-stranded circular DNA of the phage ΦX174. Their nucleotide sequences were complementary to the template, except for one nucleobase designated to introduce a nonsense mutation (Figure 1).
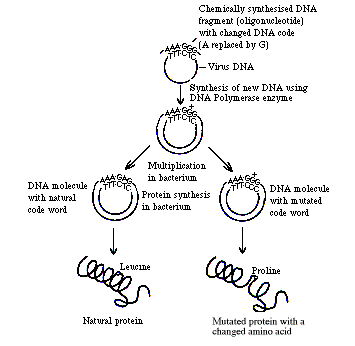
© The Royal Swedish Academy of Sciences. Source: https://www.nobelprize.org/
Figure 1: The principle of site-directed mutagenesis
The mutagenic oligonucleotides annealed the DNA template and served as a primer for DNA replication in the presence of the enzyme DNA polymerase.
The resulting product from DNA replication was a double-stranded DNA molecule that contained the point mutation derived from the mutagenic oligonucleotides.
The double-stranded DNA was circularized using the enzyme DNA ligase, separated from the residual template, and introduced into a suitable host organism for multiplication, selection, and phenotypic observation.
Reportedly, the method efficiency depended on removing non-mutagenized molecules and incomplete DNA duplexes before introducing them into the host organisms for multiplication.
The principle used in the mutagenesis of a circular single-stranded DNA template for introducing a predefined point mutation can be applied, with some modifications, to introduce deletion, insertion, and point mutations to circular and linear double-stranded DNA templates.
Nonetheless, the earlier site-directed mutagenesis methods were low in efficiency, laborious, and time-consuming. To increase the method efficiency, researchers focused on the strategy used in selecting the mutated DNA molecules and removing those that were not mutated.
The invention of polymerase chain reaction in the late 80s has enabled researchers to address the efficiency of this technique using another approach. Instead of focusing on the post-mutagenesis process, researchers can shift the focus directly to the mutagenesis process.
PCR is an in vitro technique that creates multiple copies of a designated region on the DNA template. It uses a pair of oligonucleotides as primers to bind with the template on opposite DNA strands and deoxynucleic triphosphates as building blocks to synthesize new DNA strands in the presence of a thermostable DNA polymerase enzyme. The newly synthesized DNA strands serve as templates in the following PCR rounds, thus the term chain reactions.
PCR consists of three steps: denaturation, annealing, and extension. Each step is performed only for a few seconds at different temperatures after the other in a cycle.
Theoretically, the number of the targeted DNA fragments increases twofold after the three steps are complete. After several PCR cycles, 2number of cycles PCR products are produced from the PCR reaction.
PCR is integrated into site-directed mutagenesis via the use of oligonucleotides containing the designated mutations. Only one or both PCR primers act as mutagens for site-directed mutagenesis.
The following reactions take place during PCR-based site-directed mutagenesis:
Note: In this PCR-based method, the number of mutant DNA molecules is increased exponentially after each PCR cycle. When sufficient amounts of mutant DNA are generated, the mutant DNA can be isolated from the original template either by gel electrophoresis or by restriction digestion.
Afterward, the mutant DNA can be cloned and introduced into a suitable host organism for multiplication and phenotypic observation.
Incorporating PCR into site-directed mutagenesis remarkably improved its efficiency. It simplified the separation of mutant DNA from the non-mutant template and reduced the time and labor required.
Thus, PCR-based site-directed mutagenesis is nowadays synonymous with the original mutagenesis method. To date, several variations of PCR for site-directed mutagenesis have been developed. The most notable ones are:
Overlap extension PCR involves two rounds of PCR and at least two primer pairs with one of the pair annealed to the template at the position flanking the other called nested primers.
The designated mutations are introduced in the first round of PCR reaction, using two primer pairs. Each pair consists of one mutagenic primer and one flanking primer and binds to the opposite DNA strand.
The mutagenic primers overlap the other. The flanking primers are situated at either end of the mutagenic primer-binding sites, determining the boundary of the desired DNA fragment.
In the first round, the mutagenic primers and their corresponding flanking primers anneal to the template, amplifying overlapping PCR products that contain the mutated nucleobases.
In the second round, the flanking primers anneal to the PCR product from the first round, fusing and extending the two overlapping PCR products, resulting in mutant DNA molecules of the desired length that can be multiplied and used in genetic engineering.
Overlap extension PCR can also be modified. In one modification, both PCR rounds have a non-mutagenic primer in common. In the first round, one mutagenic primer and one flanking primer bind to the template, generating PCR products that possess the designated mutations.
In the second round, the PCR products obtained from the first round act as one of the flanking primers, the so-called mega-primer, extending the fragments to meet the desired length.
The mutant DNA can be isolated using gel electrophoresis if the original template is a linear DNA. If the template is a bacterial plasmid, it can be digested with the restriction enzyme DpnI, which only cleaves methylated nucleobases absent in the PCR products.
Inverse PCR is used for mutating a circular double-stranded DNA template, using a pair of primers, each binding to the template back-to-back on the opposing strand. Both primers are mutagenic, containing the designated mutations.
Similar to overlap-extension PCR, the mutant products obtained can be isolated by digesting the plasmid template with DpnI. Then the isolated products are circularized using DNA ligase and introduced to suitable host organisms for multiplication.
This mutagenesis technique changed the way scientists study genes, proteins, their functions, and interactions. Combined with other techniques such as PCR and molecular cloning, it opens up potential technological development. For instance:
Site-directed mutagenesis has given scientists a tool to precisely manipulate the gene of interest. It enables scientists to prove the hypothesized gene function and find the underlying causes with one nucleobase precision.
Together with genetic engineering, it shortens the amount of time spent on a genetic complementation analysis and enables scientists to perform a genetic rescue experiment for lethal genes, which is not achievable using conventional breeding methods.
Such experiments are essential in determining the function, association, and inheritance of a gene or group of genes, which is fundamental in developing new approaches in genetic disease diagnosis and gene therapy.
The introduction of a precise mutation by this form of mutagenesis not only affects the genes but also the encoding proteins. Protein’s fundamental characteristics, such as their 3-dimensional structures, activities, and interaction with other molecules, are directly affected if the mutations are in the coding sequences.
On the flip side, it allows researchers to precisely engineer proteins to possess desirable characteristics. With the invention of site-directed mutagenesis, scientists finally came across the means to rationally design proteins for their intended use.
Later on, protein design expanded to include several applications such as enzyme engineering, antigen design, and protein resurfacing.
All in all, site-directed mutagenesis is a method that allows the precise modification of a gene and protein function. The integration of PCR simplifies the procedure and greatly enhances its success rate.
Along with PCR and molecular cloning, its development was revolutionary. It is one of the most fundamental techniques in molecular biology and biochemistry that opens up new areas of science and expedites technological advancement.
