

Gene shuffling is the process of creating genetic recombination in any biological system, which results in new genetic combinations that lead to differences in the genetic makeup of an individual in the population.
The unique genetic composition accounts for variations that explain the differences between individuals and population groups. Also, the diversity in the genetic makeup contributes to evolutionary forces that create novel genes and beneficial traits, which shape the population’s genetic structure over time.
Gene shuffling involves exchanging genetic elements inherited from different individuals or organisms, which exist in many forms across the three domains of life. The process gives rise to recombinants possessing novel genetic recombination.[1]
Moreover, homologous DNA segments swap in bits and pieces during the shuffling process, altering the inherent genetic information. The alteration in the DNA sequence could result in new alleles or novel combinations of different genes. Ultimately, gene shuffling can lead to characteristics or phenotypes different from the parents or donor organisms.[1-3]
Gene shuffling occurs during the sexual reproduction process of unicellular and multicellular eukaryotes. Briefly, sexual reproduction involves generating haploid reproductive cells, called gametes, from a diploid mother cell.
The haploid male and female gametes fuse in fertilization, restoring the diploid stage in the zygote. In multicellular eukaryotes, the zygote develops into an embryo, maturing and eventually becoming a new adult.[1]
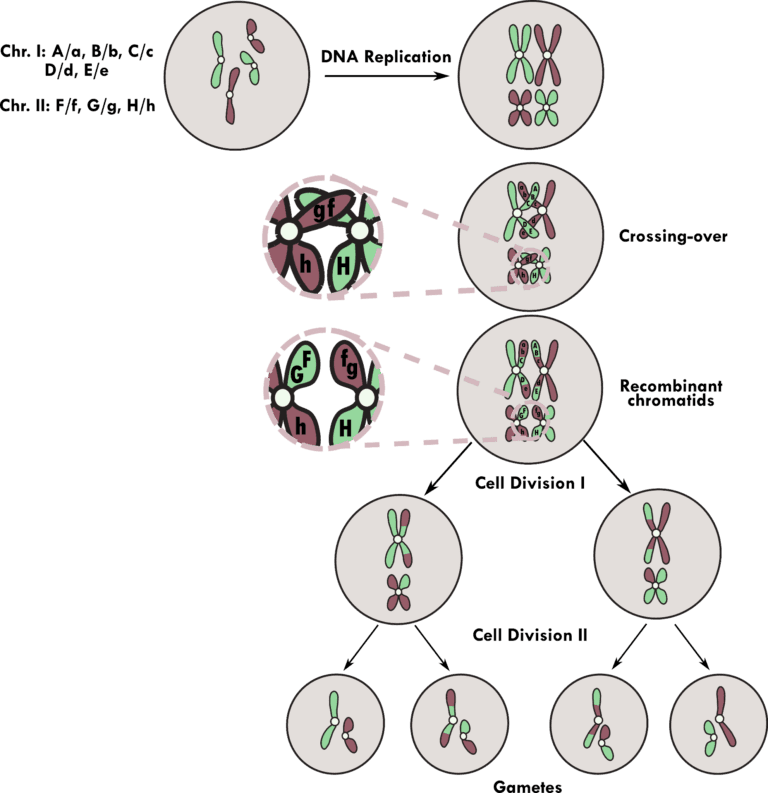
Figure 1: Reproductive cells, or gametes, are produced by meiosis in eukaryotes. Before meiosis, the mother cell possesses two sets of chromosomes inherited from both parents’ sides, as depicted in green and purple. In this case, each parent contributes different alleles, as depicted in uppercase and lowercase letters.
Meiosis begins with DNA replication in a mother cell, doubling the DNA content. After DNA replication, homologous chromosomes pair up, and gene shuffling takes place between the non-sister chromatids during a genetic recombination event termed crossing-over.
Each locus on the chromosomes independently segregate (independent assortment), except, in this case, the loci F/g and G/g, which are physically close and segregated as a linkage group.
The cell containing recombinant chromatids undergoes two rounds of cell division after genetic recombination, which results in genetically distinct gametes that possess only one set of chromosomes.
Gametes are haploid reproductive cells that possess a different genetic makeup from the originating mother cell. They are the product of meiosis, the process of nuclear and cell division that reduces the genetic material content of the originating cell in half.
At the start of meiosis, DNA molecules in a diploid mother cell undergo replication, doubling the content of the genetic material and the number of chromosomes. When DNA replication is complete, homologous chromosomes and their duplicates pair up.
The double-stranded DNA that makes up each chromosome breaks to allow the shuffling of homologous DNA segments between the paired chromosomes. Subsequently, the cell divides twice, giving rise to four daughter cells, each possessing a unique genetic combination and half of the genetic material of the mother cell.[2]
Each daughter cell is a gamete, which represents a unique recombinant. The uniqueness in the gamete genetic makeup is produced using the following recombination mechanisms:[1,4]
This recombination mechanism refers to one of Mendel’s Laws of Heredity, the Law of Independent Assortment. It states that genes governing unrelated traits are sorted independently from one another.
Independent assortment occurs during meiosis in gamete production. Alleles from the maternal and paternal sides of the mother cell have an equal chance of being shuffled into a gene locus without the influence of the other loci. This phenomenon numerically translates into an overall recombination frequency of 50%.
In other words, every gamete possesses a unique combination of various alleles in its genetic makeup. Half of the alleles are inherited from the maternal side of the mother cell, while the other half is from the paternal side.
Crossing-over is an intrachromosomal gene shuffling mechanism of homologous chromosomes. It is a remarkably precise process, which takes place during meiosis and results in recombinants, each having a different genotype.
After the duplicated homologous chromosomes align during meiosis, two chromatids break off at the same position. The broken fragments shuffle to join the homologous chromosome at the same site, replacing the fragmented chromatids. If crossing-over occurs in non-sister chromatids, the gametes will contain genetic combinations different from the parent’s.[2]
The likelihood of crossing-over is equal for any chromatid pair. This means that crossing-over occurs randomly in the genome and that in the same meiotic event, one crossing-over event has no influence on the crossing-over of the other.[1]
Nevertheless, genes located closely on the same chromosome are unlikely to assort independently. Specifically, these genes are in a genetic linkage and are shuffled and inherited as a linkage group.
The smaller the physical distance between the genes, the lower the chance they will segregate during crossing-over. Consequently, the parental allele combination is preserved in the gametes, and the recombinant frequency for these linked genes is significantly lower than 50%.[1,4]
Unlike eukaryotes, archaea and prokaryotes are single-cell microorganisms incapable of sexual reproduction. Archaea are primitive prokaryotes that inhabit extreme environments such as hot springs and deep-sea vents where the oxygen level is low, but prokaryotic bacteria are prevalent in almost all kinds of territory.
However, neither organisms can sexually reproduce nor rely on meiotic cell division to create genetic diversity.[3]
Despite the lack of sexual reproduction, gene shuffling plays a significant role in the genetic diversity of archaea and prokaryotes.
Both organisms acquire genetic diversity from horizontal gene transfer (also referred to as lateral gene transfer), which describes the exchange of genetic materials that transpire between microorganisms of the same or different species.[3,5]
In archaea, some genera of archaea bacteria can exchange their genetic materials during a fusion-like mating process. When two different archaea cells come into contact, they form a bridge that allows the two cells to fuse into one hetero-diploid cell.
The fusion of the two cells results in a hetero-diploid cell that contains two circular chromosomes and plasmids, which are genetic materials that originate from both cells.
In this stage, gene shuffling occurs in the hetero-diploid cell when parts of the two chromosomes exchange. Then, the hetero-diploid cell divides and gives rise to two daughter cells, each containing its unique rearrangement of the genetic information different from either originating cell.[5]
Gene shuffling in prokaryotes can occur following horizontal gene transfer, by which bacteria acquire novel genes and genetic elements.
There are three mechanisms of horizontal gene transfer described in prokaryotes:[6]
Unlike archaea, it is ambiguous which stage and horizontal gene transfer mechanisms involve gene shuffling.
Nonetheless, whole-genome analyses and functional studies have shown that prokaryotes shuffle the genes or genetic elements acquired from horizontal gene transfer within their genome. It builds upon the genetic diversity that prokaryotes have obtained and gives them the “flexibility” to rearrange and modify their genetics so that they can rapidly adapt to their surrounding.[7]
The direct consequence of genetic shuffling is the diversity in the genetic composition of gametes, which arises from a single set of genetic information in the parent cells.[4,7]
It allows eukaryotes to use sexual reproduction to the fullest potential. Gametes generated from meiotic cell division in eukaryotes are diverse in their genetic makeup even though they originate from the same parent.
The fusion of gametes from the opposite mating type during sexual reproduction adds variations to the genetic composition of the resulting progenies, enabling them to display different characteristics.[4]
Also termed in vitro recombination, DNA shuffling refers to a molecular biology approach created for molecular evolution. It imitates the natural gene shuffling process in vitro, which aims to generate a library of chimeric DNA molecules, producing as many versions of the genes and proteins available for choosing.
DNA shuffling begins with the fragmentation of the genes or elements of interest from various sources. The fragmentation is initially achieved by the restriction enzyme DNase I, which randomly cleaves DNA molecules.
DNA fragments are then reassembled into the original length using repeating annealing cycles and DNA polymerization using DNA polymerase. During reassembling, in vitro recombination or crossing-over happens in a template switch when the fragments anneal to their non-originating template, generating a library of chimeric DNA strands.
The obtained library is subsequently tested to select the most desirable version using a genetic engineering approach. Most DNA shuffling projects aim to create designer enzymes, a type of artificial enzyme.[11]
The location of a gene or a genetic element relative to others can advance our understanding of the inheritance — how the genes and the associated traits and characteristics pass along to the next generation.
A genetic distance is used to pinpoint a location of a gene relative to other genes or genetic landmarks on a chromosome. It uses the history of how the associated traits appear over the generations in a family to calculate a percentage of recombinant chromosomes.
One percent recombinant chromosome equals one map unit or centimorgan (abbreviated cM), which translates into a 1% chance that the gene of interest will segregate from the other gene or genetic marker due to crossing-over.[6]
Modern breeding uses pedigree information, together with the concept of genetic recombination and gene shuffling, to shorten or enhance a breeding program.
A pedigree record provides information on the occurrence of phenotypes (traits or appearance of an individual) from one generation to the next. It could suggest how the traits of interest are inherited, whether independently segregated or inherited as a linkage group.
With genotypic information, for example, the presence or absence of specific genetic markers, pedigree information is applied to select for parental combination so that the majority of the acquired offspring from the breeding program will display the most desirable traits and characteristics.[12]
Gene shuffling is a process that recombines existing genes and genetic elements in the genome to create novel genetic combinations, from which novel genes or non-existing functions can arise.
In eukaryotes, gene shuffling produces gametes for sexual reproduction, each unique in its genetic makeup. In asexually reproducing microorganisms, gene shuffling contributes to genetic diversity acquired from horizontal gene transfer.
Overall, gene shuffling is fundamental to the genetic diversity that brings about adaptive evolution and ensures species survival under the changing selective pressures. The concept of gene shuffling can advance our understanding of genes and their inheritance and applies to molecular evolution studies and modern breeding programs.
