
As an Amazon Associate, ConductScience Inc. earns revenue from qualifying purchases
Cytogenetics is the study of the structural and functional properties of the chromosome. It includes chromosomal structure, function, behavior – during mitosis and meiosis, and influence on the phenotype of an organism. It is used as a tool to understand genetic phenomena due to chromosomal aberrations. It combines two branches of biology: Cytology (which deals with cell biology); and Genetics (which deals with genes and heredity).
At the beginning of 1900, Sutton introduced the terms cytogenetics – referring to the study of the chromosome. Initially, it was used to determine the number of chromosomes present in a human being, and it took over 30 years to get the correct number[3]. Cytogenetics has evolved, through these years, from the use of simple stains to the use of digital fluorescence microscopy, and high-resolution image analysis to study the detailed structure and location of the genes and chromosomes[2]. It was an essential tool used to construct genetic maps and sequence the human genome, during the Human Genome Project.
Do You Know?★ Walther Flemming was the first person who demonstrated the human chromosome in 1882. ★ The first three chromosomal syndromes reported were Down’s syndrome, Turner’s syndrome, and Klinefelter syndrome. |
Now, with the advancement of technologies, new branches of cytogenetics have been developed; it includes molecular cytogenetics, clinical cytogenetics, and cancer cytogenetics. This article will give a brief description of clinical cytogenetics and molecular cytogenetics; and their application in clinical biology. But first, let’s learn the essential and commonly used techniques of cytogenetics.
The constant evolution of Cytogenetics, over 100 years, has introduced many essential tools for chromosomal analysis, mainly for the study of chromosomal disorders. The cytogenetic techniques[7] include:
A brief description of some of these techniques is as follows:
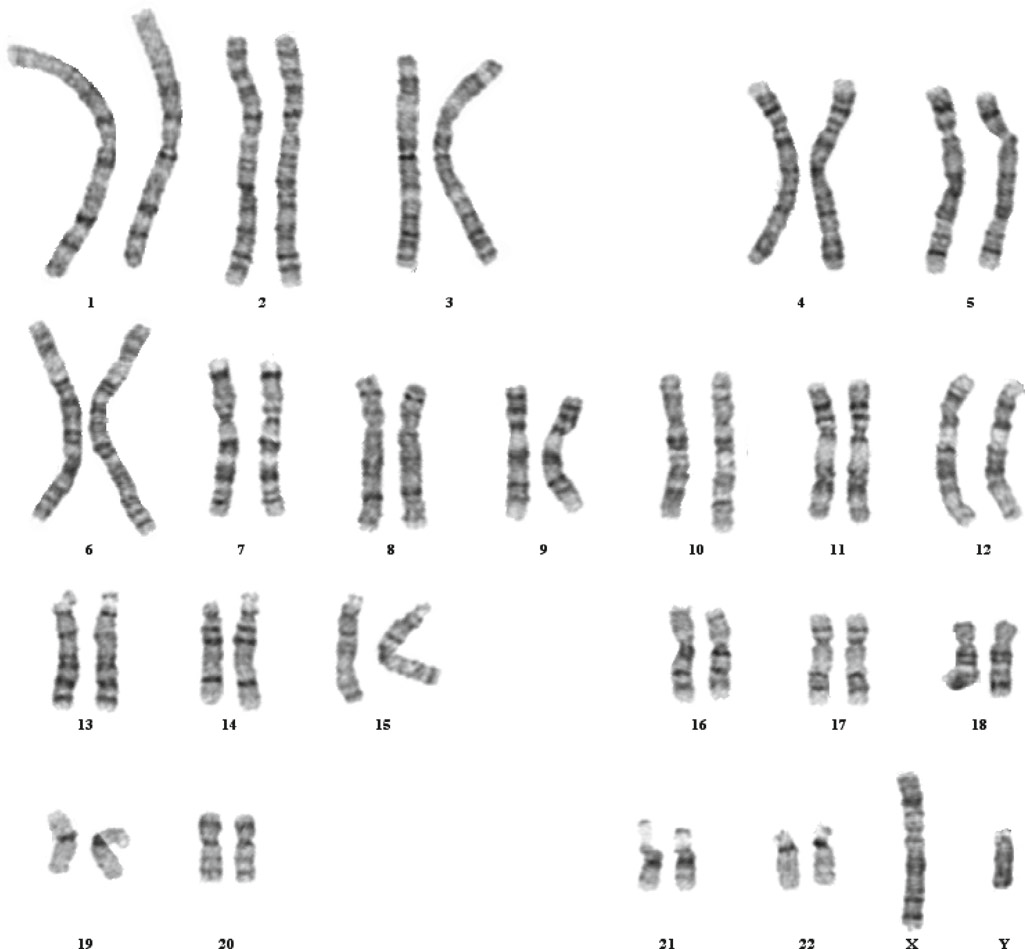
It is a process of staining chromosomes to analyze the chromosome complement of an organism, including the structure and number of chromosomes. The chromosome staining is done by banding technique, which helps to visualize the dark (gene-rich) and light (gene-poor) regions of the chromosome. The chromosomes at the metaphase (completely condensed) are arrested using colchicine and are used for the staining. This process can also be used to look for any chromosomal aberrations such as deletion, duplication, inversion, and translocation.
Banding techniques and stains[4] that are used for karyotyping and chromosome analysis are given in the table below:
| Banding technique | Stain | Staining region | Application |
| Q-banding | Quinacrine mustard or Quinacrine dihydrochloride | Brightly stains AT-rich region. | To identify the centromeric region of chromosome 3, 4, 13, acrocentric chromosome, and the Y chromosome. |
| G-banding | Giemsa stain | Darkly stains AT-rich region | Mostly used for Karyotyping. |
| R-banding | Heat in phosphate buffer followed by staining with Giemsa stain. | Brightly stains gene-rich region. | Identify gene-rich regions of the chromosome. |
| C-banding | Giemsa stain with pretreatment of barium hydroxide | Centromeric region. And the long arm of the Y-chromosome. | To identify constitutive heterochromatin region and dicentric chromosomes. |
Figure: G-banded Karyotype of a normal male. Source: Steven L. Gersen and Martha B. Keagle (2013). The principle of Clinical Cytogenetics. DOI 10.1007/978-1-4419-1688-4 |
This method was developed by Pinkel et al. in 1986 to visualize chromosomes using fluorescent-labeled probes, known as Fluorescent in situ hybridization. This technique was first used to map the human genome and to localize a specific gene on a chromosome. It is profusely used in the clinical field to detect chromosomal aberrations such as duplication, deletion, balanced/unbalanced translocations, and monosomy/trisomy. This technique is also used to study quantitative differences (copy number variation)[6]. The basic steps[7] involved in this technique are as follows:
FISH is being used by researchers in combination with other high throughput techniques to explore the uncovered mysteries of chromosomes and related diseases.
Researchers developed this cytogenetic technique to identify chromosomal regions associated with copy number variation (gains or losses of genetic information) in the genome of an organism. This method is more advanced than the FISH technique and helps to differentiate two DNA samples (test and control). The basic steps[1] involved in this method are as follows:
Although the CGH technique is very efficient, it’s very time-consuming and requires the slide preparation of metaphase chromosomes, which limits its resolution. To overcome these limitations, Scientists developed an array of CGH techniques. Array CGH is a microchip technology in which multiple reactions can be performed more precisely and in very less time.
Having discussed these Cytogenetics techniques and their clinical application, the following questions arise; “How are chromosomes examined and analyzed in the labs to detect any abnormality? What are the processes and required lab conditions?
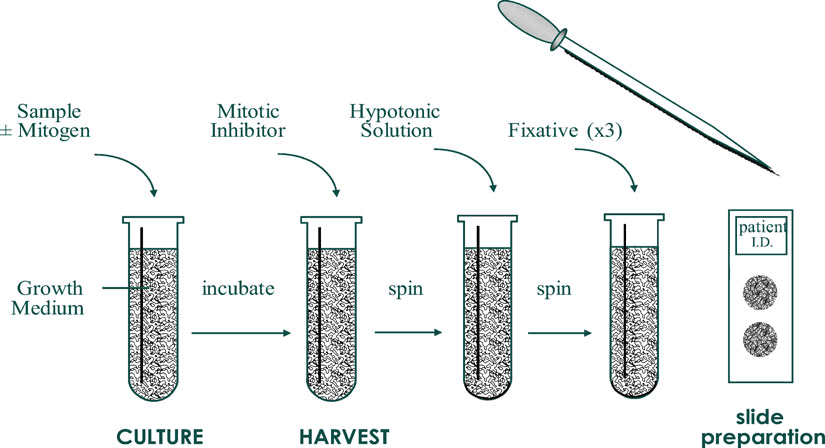
The chromosomal examination involves five major steps,[3] which are:
The source of specimen choice depends on the clinical indications and diagnosis conditions (prenatal or postnatal). But, generally, rapidly dividing cells are taken for the chromosomal analysis; to obtain the chromosomes at the metaphase stage. The specimen having spontaneously dividing cells includes two types of sources:
Researchers require the cultured cells when natural sources are inadequate to perform the analysis. The collected specimens are cultured and maintained in an aqueous growth media. Commercially, the growth media is available in two forms: complete media (ready to use solution) and incomplete media (one or more nutrients and additives are absent).
The incomplete media can be used to culture the cells after adding supplements, which include L-Glutamine, serum, antibiotics (e.g. Penicillin & kanamycin), mitogens (e.g. phytohemagglutinin), and growth factors.
After the cells are cultured, they are maintained in an open or closed system. To maintain the growth of cells, specific conditions of temperature, humidity, and pH are required in these systems. These conditions differ depending on the system in which cells are grown.
Open System:
Closed System:
When an adequate amount of cells have grown, they are harvested from the culture to perform the next step. Harvesting of cultured cells[3] includes:
After slide preparation, keep the slide at 60 °C overnight or at 90 °C for 1 hour. This is done to enhance the banding pattern and color in chromosomes.
In this stage, cells are stained by banding technique or FISH depending on the region to be highlighted or studied.
We’ve understood the process of chromosome analysis. Before we move on to the biological applications of cytogenetics, the following questions must first be addressed; “What are chromosomal aberrations? What causes it? What is their clinical significance?
Chromosomal aberrations are of two types: Structural and Numerical. Structural aberration includes Deletions, inversions, duplication, translocation, rings, and isochromosomes. Whereas, Numerical aberration involves aneuploidy and polyploidy.
The structural aberrations and their associated diseases are given in the table below:
| Structural aberration | Definition | Pathogenesis |
| Deletion | Loss of a chromosome segment | Cystic fibrosis – deletion of CFTR gene at chromosome 7 Williams syndrome – deletion of q11.23 region of chromosome 7. |
| Duplication | More than two copies of a chromosome segment are present in the same nucleus. | MeCP2duplication syndrome: duplication of the MeCP2 gene. |
| Inversion | The reverse of the normal gene order in a chromosome. | Walker-Warburg syndrome: inversion of chromosome 7 homologs. |
| Translocation | Integration of a chromosome segment with a non-homologous chromosome. | Acute myelogenous leukemia (AML): translocation of the short arm of chromosome 15 and 17. |
Numerical aberration is an alteration in the chromosome number that leads to various abnormalities in an organism. Aneuploidy is the gain or loss of a chromosome, whereas polyploidy is the gain of a set of chromosomes by a normal diploid (2n) cell. These conditions arise due to non-disjunction of the chromosomes during Meiosis I or Meiosis II.
Two common types of aneuploidy are:
Polyploidy is very common in the case of plants. But in humans, triploidy has been only observed in the case of miscarriages and polyspermy.
Cytogenetic tools are used for research and clinical purposes to identify various genetic disorders. Few examples of the application of cytogenetic techniques[5] are given below:
Molecular cytogenetics was evolved in the 1980s to analyze chromosomal aberrations based on in situ hybridization technique. The first technique that was developed was FISH (Fluorescent in situ hybridization). With the evolution of technologies, various probes are developed and FISH has been modified in various ways to attain results with very high resolutions. Some of the evolved techniques of molecular cytogenetics include: primed in situ labeling (PRINS), comparative genomic hybridization (CGH), chromosome microdissection, spectral karyotyping (SKY), Multiple color FISH (M-FISH), and array CGH. These techniques provide a close look at the chromosome structure and abnormalities.
A few ways[5] in which these techniques can be beneficial for use are explained below:
Learn more about molecular cytogenetics.
Cytogenetic techniques are, so far, the only techniques developed to analyze and characterize chromosomes. These techniques have played an essential role to map and sequence the human genome and brought a new era of coloring chromosomes. Also, they are widely used in clinical laboratories to detect chromosomal abnormalities (such as structural and numerical aberrations) during prenatal and postnatal examinations. Moreover, FISH and CGH have provided an unprecedented view of chromosomes and causes of genetic disorders. Cytogenetics is evolving and expanding (in the form of molecular and clinical cytogenetics technology) and on the way to revealing the unseen mysteries of chromosomes.

Monday – Friday
9 AM – 5 PM EST
DISCLAIMER: ConductScience and affiliate products are NOT designed for human consumption, testing, or clinical utilization. They are designed for pre-clinical utilization only. Customers purchasing apparatus for the purposes of scientific research or veterinary care affirm adherence to applicable regulatory bodies for the country in which their research or care is conducted.